1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
| import re
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
from matplotlib.collections import PatchCollection
from pymatgen.core.periodic_table import Element
class periodic_table(object):
"""
frame: str or dict, describe shapes of element cells.
str: patch class name under matplotlib.patches.
dict: {
"shape": patch class name,
"parms": width, height and other parms, needed for "shape",
"kparms": kparms, supported by "shape"
}
colors: dict, custom frame and background colors for elements.
{
"frame": str, list
"background": str, list, {"depend_on": data_key [, "cmp": color map, "cbar": True]}
}
if you use a gradient, color will based on the data from Element().data or you provide.
depend_on specify which data you want to use.
label: dict or function(not supported yet).
dict: {
"label1": [element symbols or atomic indexes],
...
}
or more detail:
dict: {
"label1": {
"elements": [element symbols or atomic indexes],
"color": not necessary
}
...
}
function: func(element symbol or atomic index) returns color and label
text: text content and format within element cells (not supported yet).
data: data beside pymatgen or you want overide for pymatgen
{symbol: {k1: v1, k2:v2}, ...}
"""
def __init__(self,
frame=None,
colors=None,
labels=None,
text=None,
data=None):
self.frame = frame or {}
if isinstance(self.frame, str):
self.frame = {"shape": self.frame or None}
self.frame.setdefault("shape", "Rectangle")
self.frame.setdefault("parms_orig", self.frame.get("parms", [1.0, 1.0]))
self.frame.setdefault("kparms", {})
self.colors = colors or {}
self.colors.setdefault("frame", "black")
self.colors.setdefault("background", "none")
if (not labels) or "background" in labels or "frame" in labels:
self.labels = labels or {}
else:
self.labels = {"background": labels}
self._text = text or None
self._data = {
Element.from_Z(i).symbol: {"Symbol": Element.from_Z(i).symbol, **Element.from_Z(i).data}
for i in range(1, 104)
}
if data:
[self._data[k].update(data[k]) for k in data]
def get_frame(self):
self.frame["parms"] = list(self.frame["parms_orig"])
if self.frame["shape"] == "FancyBboxPatch":
self.frame["kparms"].setdefault("boxstyle", "round, pad=0.1")
pad = float([*re.findall("pad=(\d+[.]\d+)", self.frame["kparms"]['boxstyle']), 0.3][0])
self.frame["parms"][0:2] = list(np.array(self.frame["parms_orig"][0:2]) - 2*pad)
try:
return getattr(mpatches, self.frame["shape"])
except AttributeError:
raise ValueError("Unsupported shape error")
def get_xys(self):
width_and_height = np.array(self.frame["parms_orig"][0:2])
text_shift = self.frame.get("text_shift",
np.array([0.5, 0.5]) * width_and_height)
if self.frame["shape"] == "FancyBboxPatch":
text_shift -= 0.10 * width_and_height
elements = [Element.from_Z(i) for i in range(1, 104)]
group_and_rows = [np.array([el.group, el.row]) for el in elements]
frame_xys = [
np.array([1, -1]) * gr * width_and_height for gr in group_and_rows
]
text_xys = [text_shift + fxy for fxy in frame_xys]
return frame_xys, text_xys
def get_colors_and_legend(self):
for v in self.colors.values():
if isinstance(v, list) and len(v) != len(self._data):
raise ValueError(
"Length of color list should be compatible with Elements.")
default_color_cycle = plt.rcParams['axes.prop_cycle'].by_key()['color']
default_edgecolor = ["black", self.colors["frame"]
][isinstance(self.colors["frame"], str)]
default_facecolor = ["none", self.colors["background"]
][isinstance(self.colors["background"], str)]
colors = {
"edgecolors":
self.colors.get("frame", default_edgecolor),
"facecolors":
[self.colors.get("background", default_facecolor),
None][isinstance(self.colors["background"], dict)]
}
legend_handles = []
for k, v in colors.items():
labels = self.labels.get({
"edgecolors": "frame",
"facecolors": "background"
}[k], None)
if labels:
if not v:
raise ValueError(
"Gradient mapped background can't be labelled.")
if isinstance(colors[k], str):
colors[k] = [colors[k]] * len(self._data)
if isinstance(list(labels.values())[0], list):
labels = {
lk: {
"elements": lv
}
for lk, lv in labels.items()
}
for i, l in enumerate(labels):
label = labels[l]
label.setdefault("label", l)
indexes = [Element(s).Z - 1 for s in label["elements"]]
label.setdefault("color", default_color_cycle[i])
for j in indexes:
colors[k][j] = label["color"]
label.setdefault("frame_kparms", {})
for lkk in ["edgecolor", "facecolor"]:
label["frame_kparms"].setdefault(lkk, "none")
label["frame_kparms"][k[0:-1]] = label["color"]
patch_legend = mpatches.Patch(
label=label["label"], **label["frame_kparms"])
legend_handles.append(patch_legend)
return colors, legend_handles
def get_plot(self, title=None, figsize=None, legend_kparms={}):
fig, ax = plt.subplots(figsize=figsize)
frame_xys, text_xys = self.get_xys()
colors, legend_handles = self.get_colors_and_legend()
frame = self.get_frame()
frames = [
frame(xy, *self.frame["parms"], **self.frame["kparms"])
for xy in frame_xys
]
collection = PatchCollection(frames, **colors)
ax.add_collection(collection)
[
ax.text(
*text_xys[i],
Element.from_Z(i + 1).symbol,
horizontalalignment="center",
verticalalignment="center",
family="Times New Roman",
size="x-large"
) for i in range(len(self._data))
]
if legend_handles:
legend_kparms.update({"handles": legend_handles})
legend_kparms.setdefault("loc", "upper center")
legend_kparms.setdefault("bbox_to_anchor", (0.41, 1.0))
legend_kparms.setdefault("fontsize", "x-large")
ax.legend(**legend_kparms)
if title:
plt.title(title, fontsize=28)
plt.axis('equal')
plt.axis('off')
plt.tight_layout()
return plt
def show(self, title=None, figsize=None):
plt = self.get_plot(title=title, figsize=figsize)
plt.show()
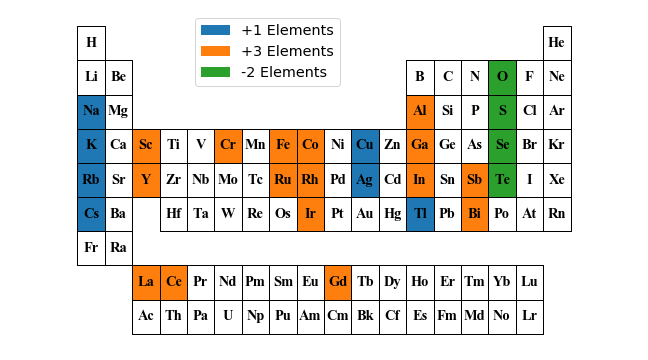
p = periodic_table(
frame={
"shape": "Rectangle",
"parms": [1.0, 1.25]
},
labels={
"background": {
"+1 Elements": ['Na', 'K', 'Rb', 'Ag', 'Cs', 'Tl', 'Cu'],
"+3 Elements": [
'Al', 'Sc', 'Cr', 'Fe', 'Co', 'Ga', 'Y', 'Ru', 'Rh', 'In',
'Sb', 'La', 'Ce', 'Gd', 'Ir', 'Bi'
],
"-2 Elements": ['O', 'S', 'Se', 'Te']
}
})
pplt = p.get_plot(figsize=(9, 5))
pplt.show()
plt.close()
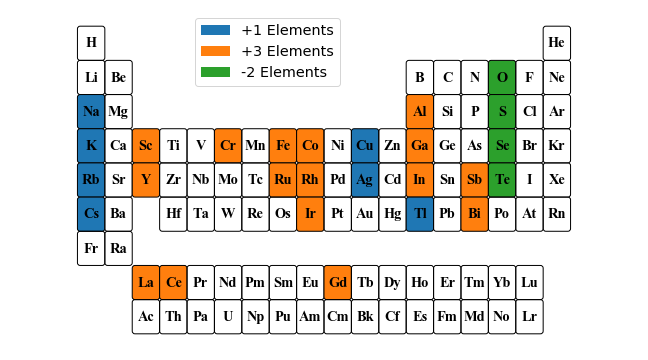
p = periodic_table(
frame={
"shape": "FancyBboxPatch",
"parms": [1.0, 1.25]
},
labels={
"background": {
"+1 Elements": ['Na', 'K', 'Rb', 'Ag', 'Cs', 'Tl', 'Cu'],
"+3 Elements": [
'Al', 'Sc', 'Cr', 'Fe', 'Co', 'Ga', 'Y', 'Ru', 'Rh', 'In',
'Sb', 'La', 'Ce', 'Gd', 'Ir', 'Bi'
],
"-2 Elements": ['O', 'S', 'Se', 'Te']
}
})
pplt = p.get_plot(figsize=(9, 5))
pplt.savefig("periodic_table.pdf", transparent=True)
pplt.show()
|